Simulated time series¶
In this example, we generate discrete time series data from a stochastic
model with known parameters. After generating the data, we demonstrate
how to use FEM to infer the model parameters.
We start by importing the necessary packages and ensuring that matplotlib places its output in the notebook:
In [1]:
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import fem
%matplotlib inline
We consider a system of \(n\) variables \(x_i(t)\) that each take on values from the first \(m\) nonnegative integers over time, i.e. \(x_i(t)\in\{0,\ldots,m-1\}\) for all \(i=1,\ldots,n\) and \(t=1,\ldots,\ell\). The state of variable \(i\) at time \(t\), \(x_i(t)\), is represented by the one-hot encoding \(\sigma_i(t)\in\{0,1\}^m\) with components
while the state of the system at time \(t\) is represented by their concatenation \(\sigma(t)=\begin{pmatrix}\sigma_1(t)&\cdots&\sigma_n(t)\end{pmatrix}^T\).
In the model that we consider, the probability of \(x_i(t+1)=j\) depends on \(\sigma(t)\) and the model parameters \(W\in\mathbb{R}^{mn\times mn}\)
according to the Boltzmann distribution.
The FEM package implements a helper function
fem.discrete.model_parameters for generating random model parameters
for the above model. In this case, we restrict each variable to taking
the same number of states as all the other variables by passing an
integer m to the function, though the function supports m an
iterable of length n that specifies the number of states per
variable. By default, the parameters in w are sampled from a normal
distribution with mean 0 and variance \(1/\sqrt{mn}\) (using
optional arguments dist=np.random.normal and
dist_par=(0, 1/np.sqrt(m*n))). The model parameters w are
returned as a dictionary with keys corresponding to degrees of the model
(degs=[1] by default, see FEM for discrete
data for more information).
In [2]:
n, m = 10, 3
w = fem.discrete.simulate.model_parameters(n, m)
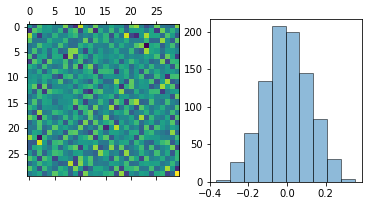
We plot w as an image and flatten out w to a 1D array and plot
the distribution:
In [3]:
fig, ax = plt.subplots(1, 2, figsize=(6,3))
ax[0].matshow(w[1])
w_flat = np.hstack([wi for wi in w.itervalues()]).flatten()
hist = ax[1].hist(w_flat, bins=n, ec='k', alpha=0.5)
Next, we generate time series data given the model parameters w. The
FEM package implements a helper function
fem.discrete.simulate.time_series for generating time series data
according to the model above. We specify with the parameter o that
the number of time points to be returned is \(o\) times the number
of model parameters (\((mn)^2\) in this case). We could instead
specify a number of time points with the optional argument l. The
columns of the returned array store the values of the system at
different times \(t\).
In [4]:
x = fem.discrete.simulate.time_series(w, n, m, o=4.0)
Let’s examine the generated data. Each variable takes m=3 unique
states as expected. By default, these are symbolized by the first
nonnegative integers. The average of each variable is approximately 1.
In [5]:
tab = []
for i in range(n):
tab.append([np.unique(x[i]), x[i].mean()])
pd.DataFrame(data=tab,
columns=['states', 'average'],
index=['$x_{%i}$' % (i,) for i in 1+np.arange(n)])
Out[5]:
| states | average | |
|---|---|---|
| $x_{1}$ | [0, 1, 2] | 1.015556 |
| $x_{2}$ | [0, 1, 2] | 1.006389 |
| $x_{3}$ | [0, 1, 2] | 0.999722 |
| $x_{4}$ | [0, 1, 2] | 1.005000 |
| $x_{5}$ | [0, 1, 2] | 0.996389 |
| $x_{6}$ | [0, 1, 2] | 1.002500 |
| $x_{7}$ | [0, 1, 2] | 0.970556 |
| $x_{8}$ | [0, 1, 2] | 0.989444 |
| $x_{9}$ | [0, 1, 2] | 0.980556 |
| $x_{10}$ | [0, 1, 2] | 1.000278 |
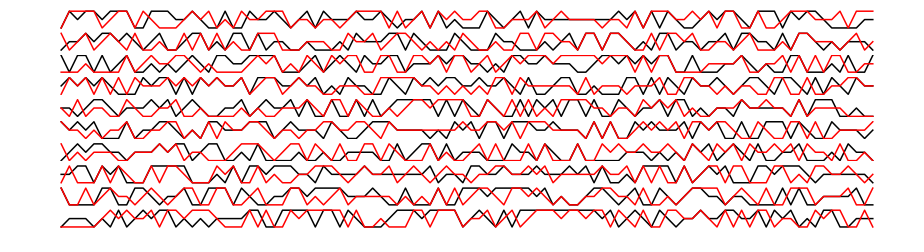
Plotting the first 100 time points, there is no obvious pattern to the naked eye:
In [6]:
fig, ax = plt.subplots(n, 1, figsize=(16,4))
for i in range(n):
ax[i].plot(x[i, :100], 'k-')
ax[i].axis('off')

We can however determine the model that generated the above data using
FEM. We start by splitting the data x into two sets x1,
which contains all but the last time point, and x2, which contains
all but the first time point. Next, we use the fem.discrete.fit.fit
function to infer the w that is used to map x1 to x2. In
this case, we require a fixed number of iterations of the method with
the iters argument.
The fit function returns the inferred parameters (w_fit below) as a
dictionary with keys degs (degs=[1] by default). It also returns
the discrepancy (disc below) for each variable defined by
.
The lists in disc are running values of \(d_i\) as \(W\) is
updated by the iteration.
In [7]:
mod = fem.discrete.fit.model()
x1, x2 = x[:, :-1], x[:, 1:]
mod.fit(x1, x2)
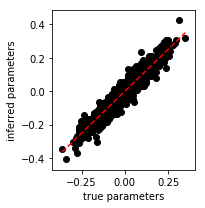
We make a scatter plot of the inferred w_fit model parameters versus
the true w model parameters. Since, the points are scattered around
the line with slope 1 that passes through the origin, we confirm that
the algorithm accurately recovers the true model parameters.
In [8]:
w_fit_flat = np.hstack([wi for wi in mod.w.itervalues()]).flatten()
fig = plt.figure(figsize=(3,3))
ax = plt.gca()
ax.scatter(w_flat, w_fit_flat, c='k')
lo, hi = w_flat.min(), w_flat.max()
grid = np.linspace(lo, hi)
ax.plot(grid, grid, 'r--')
ax.set_xlabel('true parameters')
ax.set_ylabel('inferred parameters')
plt.tight_layout()
plt.show()
In [9]:
x2_prediction, p = mod.predict(x1)
In [10]:
fig, ax = plt.subplots(n, 1, figsize=(16, 4))
for i in range(n):
ax[i].plot(x2[i, :100], 'k-')
ax[i].plot(x2_prediction[i, :100], 'r-')
ax[i].axis('off')
plt.show()